


Signaling pathways are the intricate networks that govern cell behavior, lying at the crux of cell function. They are used by nature to control almost all forms of cell activity – growth, response to stimuli, adaptation, replication, cell death. At its most fundamental, a signaling pathway is a cascade of activity, whereby a single stimulus triggers a precise sequence, resulting in specific cellular outcomes.
An analog for a signaling pathway is the board game “Mouse Trap.” In the game, once properly assembled, one single action – the turning of a crank – leads to a precisely defined series of subsequent actions – the kicking of a marble to roll down a staircase, the swinging of a seesaw, the tipping of a bucket – that ultimately results in the dropping of a cage and the capture of the mouse. In a cell, these actions range from the mechanical, like dimerization or recruitment of other protein components, to the enzymatic, like phosphorylation or ubiquitination.
While some signaling pathway components are linear and relatively straightforward, others involve tremendous complexity, relying on countless proteins to collectively transduce a signal further downstream. These orchestrations depend on the so-called “scaffolding” function of proteins to physically associate with each other in a cell. These resulting protein complexes can enable potent signal perpetuation leading to disease progression.
While the therapeutic potential of targeting signaling pathways is known and established, the majority of therapeutic approaches involve targeting one single component of a pathway, as opposed to the more complicated, though perhaps more impactful, disruption of complex formation.
Imagine you’re trying to block a car’s access to Manhattan. The traditional targeting of a signaling pathway, likened to a selective kinase inhibitor, would be like blocking access to the Brooklyn Bridge or the Holland Tunnel – undoubtedly an initially effective way to prevent a car from accessing the island. However, let’s consider a different approach. What if we were able to disable the electricity for every single intersection in Manhattan? Rather than just a single access point being blocked, the entire system would be completely hamstrung, with traffic impeded in all roads, intersections, bridges, and tunnels. A similar degree of control could be achieved in a cell if an entire signaling complex were disrupted, since the pathway’s ability to further perpetuate a downstream signal would be completely disabled.
There is clear precedent for such a complex-directed therapeutic approach in drug discovery. Rapamycin, which targets the mTORC1 complex, has been successfully used in oncology to treat advanced perivascular epithelioid cell tumors, in addition to applications in other disease areas, like organ transplantation. Inflammasome inhibitors are currently being developed for a range of autoinflammatory diseases. Myddosome inhibitors have shown promise in autoimmune disorders and hematological cancers.
One such pathway that we believe holds significant promise for a complex-directed therapeutic approach is the NF-kB pathway. NF-kB is a key mediator of inflammatory responses, playing a crucial role in the regulation of cytokine production, as well as in the activation and proliferation of immune cells. Dysregulation of this pathway is associated with a wide spectrum of diseases, including a number of cancers, such as lymphomas and certain solid tumors, and autoimmune diseases.
Within the NF-kB pathway, one crucial cog in the signaling cascade is mucosa-associated lymphoid tissue lymphoma translocation protein 1, or MALT1. MALT1 is a component of the CBM complex – the “Manhattan” of this pathway – which plays a key regulatory role in the signaling in cells, including in B and T cells.
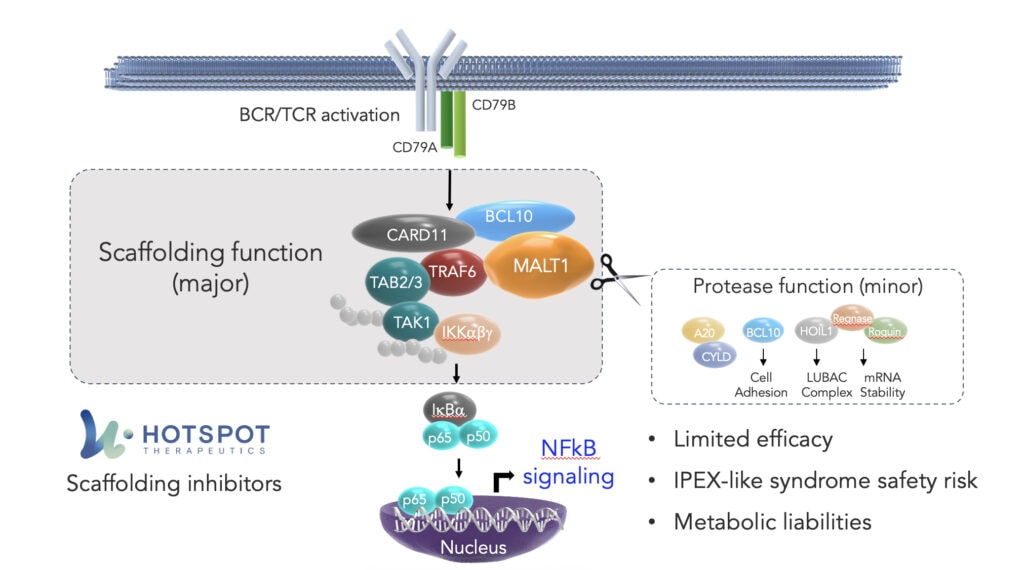
Within the clustering of the CBM complex, MALT1 has two functions: a scaffolding function, essential to the assembly of the complex, and a protease function, controlling fine tuning, which collectively result in the activation of NF-kB.
The protease function of MALT1 is being extensively targeted by the biopharmaceutical industry, most notably with Janssen’s MALT1 inhibitor, safimaltib, that demonstrated clinical proof of concept in patients with relapsed/refractory Non-Hodgkins lymphoma and chronic lymphocytic leukemia. While these early data have shown promising proof of principle for the therapeutic targeting of MALT1, these initial results suggest challenges not only with potency, but also tolerability, suggesting that a protease function-directed approach may ultimately have limitations on the candidate’s ultimate efficacy in these cancers.
At HotSpot, we believe our proprietary Smart AllosteryTM drug discovery platform affords us a unique understanding of how nature controls proteins through signaling pathways, including at the intricacies of complex formation. A precise understanding of these dual functions points to MALT1’s scaffolding function as the key node for inhibition by a therapeutic agent. Recognizing the therapeutic potential of effectively disrupting the CBM complex in the NF-kB pathway, using our technology, we have developed small molecules that we believe to be the first and only inhibitors of MALT1’s scaffolding function.
At the recent American Society of Hematology (ASH) Annual Meeting, we presented data for the first time supporting the highly differentiated nature of MALT1 scaffolding inhibition. Compared to conventional protease inhibitors, scaffolding inhibition delivered compelling potency and efficacy in in vitro and in vivo models. Moreover, scaffolding inhibitors demonstrated no deleterious effects on effector T cell functions or T-regulatory cell numbers, which we believe enables potential combination with CAR-T or T-cell engagers without any negative effects on those immune therapies.
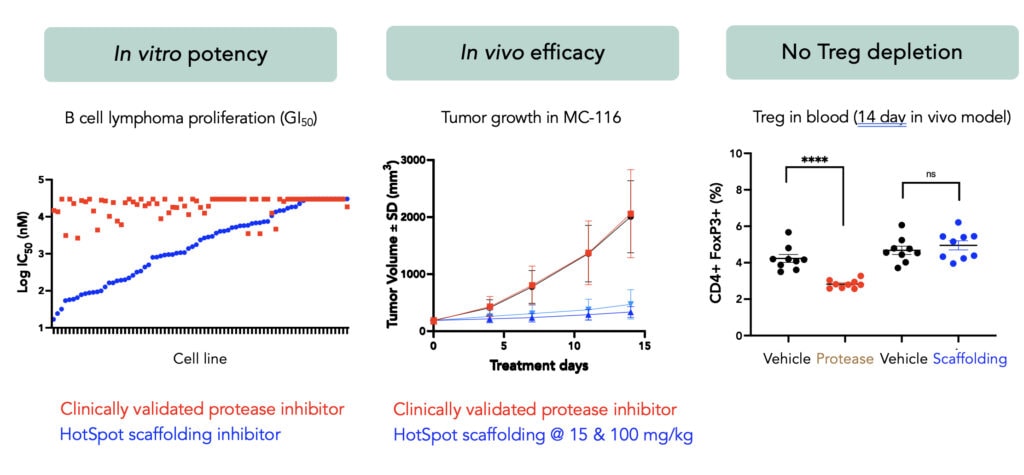
We are now focused on advancing our Development Candidate, HST-1021, into the clinic to patients. This is an exciting development both for HotSpot and for the field – not only has the CBM complex been successfully targeted in vitro and in vivo for the first time, but we believe we now have a roadmap for drugging other signaling assemblies relevant to disease.